Just when you thought you knew E. coli ... [Updated]
- Share via
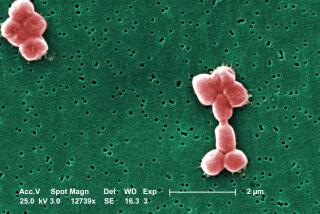
An entirely new, and multiple-drug-resistant, strain of E. coli appears to be responsible for the deadly outbreak of the bacterium in Europe, scientists announced Thursday.
Called a mutant form of two other strains of the bacterium, the new bacterium seems to be more virulent and toxin-producing than your garden variety strains of E. coli. And it appears to be giving E. coli O157:H7 a run for its money in the ability to alarm consumers. That’s the strain most of us have come to fear after several outbreaks in this country.
After a preliminary analysis of the bacterium, the Beijing Genomics Institute in China said in a news release that the “super-toxic” E. coli strain carries genes that confer resistance to several antibiotics, including aminoglycoside, macrolides and Beta-lactam antibiotics.
Antibiotic treatment has been proving ineffective against the strain, found the University Medical Centre Hamburg-Eppendorf, which has treated the majority of the infected patients in northern Germany. The Beijing institute worked with the German hospital to sequence the bacterium’s genome in three days.
According to the institute, the new E. coli strain is a close genetic relative to a strain, EAEC 55989, found in the Central African Republic. But the new strain also carries genes that appear similar to ones involved with bloody diarrhea (hemorrhagic colitis) and hemolytic-uremic syndrome, a sometimes fatal kidney disease.
The sequences of the new bacterial strain, a new type of EHEC serotype O104 E. coli strain, are available for download here, and the Beijing Genomics Institute can be followed on Twitter: @BGI_Events.
The analysis appears preliminary, notes geneticist Sophien Kamoun at the Sainsbury Laboratory in Norwich, Britain, who took a peek at the posted data. Kamoun wrote in an email:
“A genome assembly will be needed for more conclusive analyses to be conducted. ... It requires some computational analyses and could require more sequencing. I suppose they are doing it as we speak. It is unusual to have a press release about the unassembled reads, because there is absolutely no guarantee that the data generated so far will be sufficient to generate a robust genome assembly.”
The Beijing Genomics Institute said it expects to post the genome assembly in the next few days to a week.
[For the record at 8:28 a.m. June 2: An earlier version of this post stated that the Beijing Genomics Institute said it expects to post the E. coli genome assembly in the two or three days. It anticipates that posting in next few days to a week.]
RELATED: More news from HealthKey